Environmental DNA (or eDNA) profiling is being used increasingly to analyse a range of substrates and historical samples, apparently even thin air, but what about soil? Join Prof. Andy Lowe as he investigates an important and cost-effective method of environmental DNA profiling.
Monitoring the state of our environment is the only way we can be sure whether we are improving or degrading our natural and production systems and the life support systems we depend on.
Technology is increasingly playing a role in the search for cost-effective monitoring methods and proxies, including camera traps and acoustics. But are we missing an important and cost-effective method in environmental DNA profiling of soils?
Environmental DNA (or eDNA) profiling is being used increasingly to analyse a range of substrates and historical samples, apparently even thin air, but what about soil?
We all know it’s important
Soil is a complex matrix of minerals, organic matter, gas, water and living organisms and hosts a quarter of the earth’s biodiversity. Soil is a vital component of our production and natural systems, allowing the replenishment and acquisition of water, nutrients and other functions critical to the survival of plants, and of all life on earth and has been estimated to contribute between US $1.5 – 13 trillion to the value of ecosystems services every year (Laban, Metternicht & Davies 2018). In addition, recent work has highlighted the human health benefits from exposure to environmental microbiomes, including healthy immunological development (Mills et al., 2020).
But how do we monitor the soil microbiome? Traditional field-based monitoring focuses on terrestrial macro-organisms (e.g. plants, birds), vegetation regrowth (e.g. growth, biomass), or subjective classification (e.g. land condition, vegetation cover). But all of these methods are logistically demanding, hard to standardise, and largely discount the microbiome. We need monitoring methods that are quick and easy and offer a cost effective and simple way to track the condition, status and trajectory of associated ecosystems.
With advancements in computing power and DNA sequencing technology comes the capability to provide greater insights into this previously little-known biological world. High-throughput amplicon sequencing of eDNA is a cost-effective, scalable and uniform ecological monitoring solution. Little prior knowledge is needed to identify a representative suite of species, and unlike orthodox field methods, it can provide a more accurate picture of diversity, by targeting focal species groups, such as bacteria and fungi.
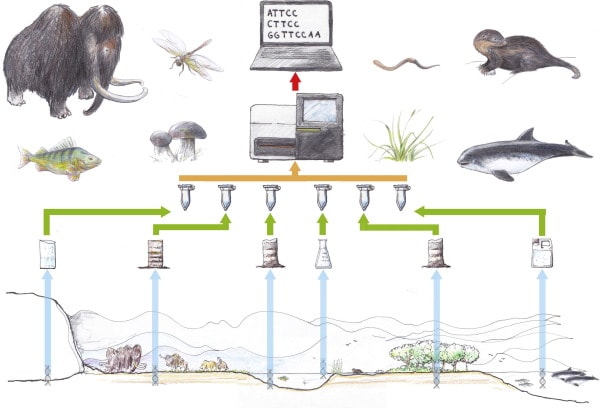
The overall workflow for environmental DNA (eDNA) studies (Thomsen et al, 2015) “Environments and their respective samplings from left to right: (i) glaciers; (ii) permafrost/tundra; (iii) aquatic sediments; (iv) lakes and streams; (v) terrestrial habitats; (vi) oceans.” Drawing by Lars Holm, published in Thomsen et al, 2015.
Setting a baseline
DNA sequence analysis has also already been widely employed to investigate the health, function and degradation of a broad range of environments and biological systems. From hot springs (Baker et al. 2001) to municipal wastewater (Miura et al. 2007), spacecraft clean rooms (Moissl et al. 2007) to systems and practices, such as agricultural land management (García-Orenes et al. 2013; Rampelotto et al. 2013), soil tillage (Ondreicková et al. 2018), vineyard management (Holland et al. 2016) and a range of land use systems and practices (Jeanbille et al. 2016).
Using these methods, a continental-scale map of the soil microbiome has been completed – the Biomes of Australian Soil Ecosystems or BASE program (Bisset et al 2016). More than 1500 sites across Australia and the Antarctic, spanning deserts, agricultural lands, the tropics, alpine regions, and coastal areas have been sampled and provide invaluable base-line data on the soil microbial community.

Soil sampling sites for the BASE program
Microbiome data can also be combined with other environmental data, including meteorological data, climate data, biological surveys, geochemical information and vegetation type. TERN long-term monitoring sites form part of the sampling effort, particularly the survey plots aligned as transects across ecosystems and TERN SuperSites, that monitor ecosystem processes. Analysis of these sites shows that turn over in environmental variation also drives microbiome turnover, which can be significant over short geographic scales.
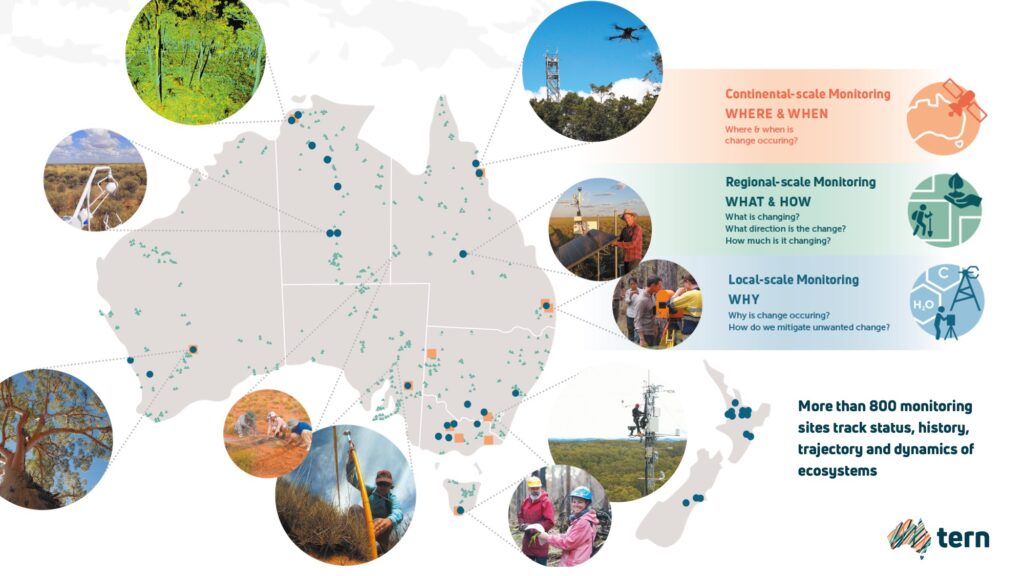
TERN’s long-term ecosystem monitoring sites form part of the national soil sampling effort
Tracking community change
Against these natural microbiome distributions, it’s possible to track how community composition changes with landuse change (e.g. Gellie et al 2017) and is an important monitoring tool to assess soil condition.
Soil can also provide a useful matrix to analyse the presence of other organisms living on and in soil, not just microbes. When animals or plants interact with their environment, they inevitably shed or deposit different types of tissues, secretions, or excrement. As a result, the DNA of fauna and flora occur naturally in the environment. Such DNA, which has accumulated in environmental samples, for example, soil, stream water or air, can be analysed using high-throughput sequencing methods, and has the potential to revolutionise and massively speed up biodiversity survey and monitoring protocols.
In a recent pilot study, soil samples maintained by TERN were analysed for their capacity to give up signatures of faunal and floral presence via eDNA analysis. Although present at low concentration both floral and faunal DNA could be recovered with high replicability and can potentially be used as a survey method.
However, it should also be noted that only a small number of sequences could be detected when analysing the vertebrate community. Therefore, a more targeted approach for sampling may improve the efficiency of obtaining data for the diversity of local fauna and flora from soil surveys.
Soil samples from more than 800 sites across the Australian continent are available for further analysis.
Summary
The soil microbiome is all-important, all-pervasive, and can certainly be used for screening and analysing the health condition and trajectory of ecosystems.
And it is not just the soil microbes that can be analysed in this way. The potential exists to also include environmental samples from other ecosystems, e.g. water, pollen. Such analyses can provide rapid, environmental monitoring protocols. This means whole ecosystems can be surveyed and monitored at a large scale at a relatively low cost, and can be linked with ecological function and condition assessment of sites.
It is easy to see then how eDNA analysis of soil is here to stay and will be an increasingly important environmental monitoring method of the future.
- Article by Andrew Lowe.
- With input and feedback from Arif Malik, Alex Mason, Nick Gellie, Jason Mills
References
Baker, G, Gaffar, S, Cowan, D, Suharto, A 2001, ‘Bacterial community analysis of Indonesian hot springs’, FEMS Microbiology Letters, vol. 200, no. 1, pp. 103-109.
Bissett, A, Fitzgerald, A, Meintjes, T, Mele, P, Reith, F, Dennis, P, Breed, M, Brown, B, Brown, M, Brugger, J, Byrne, M, Caddy-Retalic, S, Carmody, B, Coates, D, Correa, C, Ferrari, B, Gupta, V, Hamonts, K, Haslem, A, Hugenholtz, P, Karan, M, Koval, J, Lowe, A, Macdonald, S, McGrath, L, Martin, D, Morgan, M, North, K, Paungfoo-Lonhienne, C, Pendall, E, Phillips, L, Pirzl, R, Powell, J, Ragan, M, Schmidt, S, Seymour, N, Snape, I, Stephen, J, Stevens, M, Tinning, M, Williams, K, Yeoh, Y, Zammit, C, Young, A 2016, ‘Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database’, GigaScience, vol. 5, no. 1, https://doi.org/10.1186/s13742-016-0126-5
García-Orenes, F, Morugán-Coronado, A, Zornoza, R, Scow, K 2013, ‘Changes in Soil Microbial Community Structure Influenced by Agricultural Management Practices in a Mediterranean Agro-Ecosystem’, PLoS ONE, vol. 8, no. 11, https://doi.org/10.1371/journal.pone.0152958
Gellie NJC, Mills JG, Breed MF, Lowe AJ (2017) Revegetation rewilds the soil bacterial microbiome of an old field. Molecular Ecology 26(11): 2895-2904.
Holland, T, Bowen, P, Bogdanoff, C, Lowery, T, Shaposhnikova, O, Smith, S, Hart, M 2016, ‘Evaluating the diversity of soil microbial communities in vineyards relative to adjacent native ecosystems’, Applied Soil Ecology, vol. 100, pp. 91-103.
Jeanbille, M, Buée1, M, Bach, C, Cébron, A, Frey-Klett, P, Turpault, M, Uroz1, S 2016, ‘Soil Parameters Drive the Structure, Diversity and Metabolic Potentials of the Bacterial Communities Across Temperate Beech Forest Soil Sequences’, Microbial Ecology, vol. 71, pp. 482-493.
Laban, P, Metternicht, G, Davies, J 2018, ‘Soil Biodiversity and Soil Organic Carbon: keeping drylands alive’, Gland, Switzerland: IUCN, https://doi.org/10.2305/IUCN.CH.2018.03.en
Lemetre, C., Maniko, J., Charlop-Powers, Z., Sparrow, B., Lowe, A., and Brady, S. (2017) Bacterial natural product biosynthetic domain composition in soil correlates with changes in latitude on a continent wide-scale. Proceedings of the National Academy of Sciences.
Mills, J, Bissett, A, Gellie, N, Lowe, A, Selway, C, Thomas, T, Weinstein, P, Weyrich, L, Breed, M 2020, ‘Revegetation of urban green space rewilds soil microbiotas with implications for human health and urban design’, Restoration Ecology, vol. 28, pp. 322-334.
Miura, Y, Hiraiwa, M, Ito, T, Itonaga, T, Watanabe, Y, Okabe, S 2007, ‘Bacterial community structures in MBRs treating municipal wastewater: Relationship between community stability and reactor performance’, Water Research, vol 41, no. 3, pp. 627-637.
Moissl, C, Osman, S, La Duc, M, Dekas, A, Brodie, E, DeSantis, T, Venkateswaran, K 2007, ‘Molecular bacterial community analysis of clean rooms where spacecraft are assembled’, FEMS Microbiology Ecology, vol. 61, no. 3, pp. 509-521.
Ondreicková, K, Piliarová, M, Bušo, R, Hašana, R, Schreiber, L, Gubiš, J Kraicm J 2018, ‘The Structure and Diversity of Bacterial Communities in Differently Managed Soils Studied by Molecular Fingerprinting Methods’, Sustainability, vol. 10, no. 4, pp. 1095.
Rampelotto, P, de Siqueira Ferreira, A, Barboza, A, Fernando, L, Roesch, W 2013, ‘Changes in Diversity, Abundance, and Structure of Soil Bacterial Communities in Brazilian Savanna Under Different Land Use Systems’, Microbial Ecology, vol. 66, pp. 593-607.







